Describe the Process of Alternative Splicing
Alternative splicing is the process of producing variably spliced mRNAs by selecting alternative combinations of splice sites within a messenger RNA precursor pre-mRNA. The expression of a single gene can result in multiple proteins with the.

Alternative Splicing Importance And Definition Technology Networks
1 Splicing allow for a process called alternative splicing in which more than one mRNA can be made from the same geneAlternative splicing of precursor mRNA is an essential mechanism to increase the complexity of gene expression and it plays an im.
. In splicing some sections of the RNA transcript introns are removed and the remaining. A process known asalternative splicing allows for different combinations of exons to be included in the final mature mRNA making different versions of proteins called isoforms that are all encoded by the same gene. 100 1 rating Q.
This process is called RNA splicing. An alternative splicing pathway uses another set of snRNPs that comprise the U12 spliceosome. It produces a final draft of the mRNA before translation gets under way.
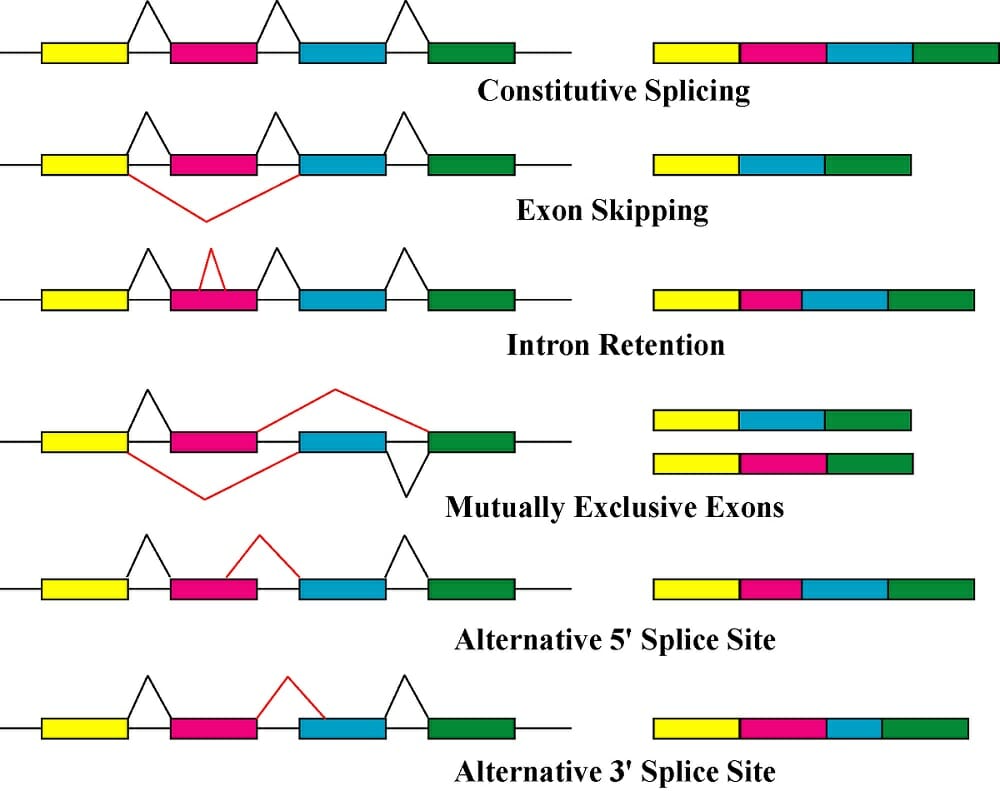
Self-Splicing and Alternative Splicing Figure 2. Alternative splicing of mRNA allows for many proteins to be made with different functions all produced from a single gene. Despite the fact that these various mRNAs all come from the same gene they can encode proteins with different sequences and activities.
Some of the genes undergo self-splicing eg. In eukaryotes alternative splicing refers to a process via which a single precursor RNA pre-RNA is transcribed into different mature RNAs. Alternative splicing is a process to differentially link exon regions in a single precursor mRNA to produce two or more different mature mRNAs a strategy frequently used by higher eukaryotic cells to increase proteome diversity andor enable additional post-transcriptional control of gene expression.
Here the introns can catalyse their own excision from their parent RNA. When an RNA transcript is first made in a eukaryotic cell it is considered a pre-mRNA and must be processed into a messenger RNA mRNA. It is called alternative splicing.
This process can take place either co-transcriptionally or post-transcriptionally. No 1 Answer a Alternative splicing is the process of splicing different subsets of exons from the same gene to produce different combinations and therefore different proteins. Works in a similar way to major intron splicing GU-AG.
A schematic representation of alternative splicing. The proteins bind to specific sites on the pre-mRNA and tell the splicing factors which exons should be used. A 5 cap is added to the beginning of the RNA transcript and a 3 poly-A tail is added to the end.
Alternative splicing is the joining of different splice sites 5 and 3 allowing individual genes to express multiple mRNAs that encode proteins with diverse and even antagonistic functions. These multiple mRNAs can encode proteins that vary in their sequence and activity and yet arise from a. Therefore alternative splicing a type of post-transcriptional modification is the process by which exons or portions of exons or non-coding regions within a pre-mRNA transcript are differentially joined or skipped resulting in multiple protein isoforms being encoded by a.
Many studies suggest that the vast majority of human genes contain introns and that most pre-mRNAs undergo alternative splicing Jutzi et al 2018. This principle is called alternative splicing. Some mitochondrial genes are also capable of self.
Instead its typically controlled by regulatory proteins. By this process one gene can. Alternative splicing is a molecular mechanism that modifies pre-mRNA constructs prior to translation.
We review their content and use your feedback to keep the quality high. Thus alternative splicing enables the translation of a limited number of coding genes into a large number of. Phage genes protozoan ribosomal RNA genes etc.
Alternative splicing or alternative RNA splicing or differential splicing is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. This process is called alternative splicing and it makes it possible to produce different proteins from the same gene these different protein versions from the same gene are called isoforms. Alternative splicing is not a random process.
In this process particular exons of a gene may be included within or excluded from the final processed messenger RNA mRNA produced from that gene. In this way our bodies can produce over 100000 proteins from only 20000 genes. This process increases the diversity of proteins and occurs as a normal splicing process in most eukaryotes.
Alternative splicing is the biological process responsible for the production of variant mRNA molecules from a particular primary RNA transcript of a particular gene. This process can produce a diversity of mRNAs from a single gene by arranging coding sequences exons from recently spliced RNA. Alternative splicing refers to the process by which a given gene is spliced into more than one.
943 Alternative Splicing Regulation. Alternative splicing is the process of selecting different combinations of splice sites within a messenger RNA precursor pre-mRNA to produce variably spliced mRNAs. Alternative splicing mRNA processing event in which some exons are removed or joined in various combinations.
The target introns are defined by longer consensus sequences at the splice junctions rather than strictly according to the GU-AG or AU-AC rules.

Alternative Splicing An Overview Sciencedirect Topics
Gene Splicing Mechanism Alternative Splicing Tutorial Splice Variant Detection

Alternative Splicing Definition Explanation Examples Biology Dictionary
No comments for "Describe the Process of Alternative Splicing"
Post a Comment